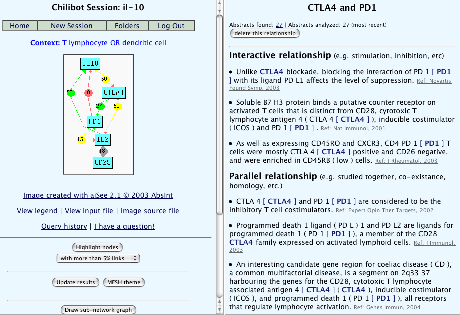
Chilibot (paper published last year in BMC Bioinformatics) is the first MEDLINE-mining tool that I've actually found to be useful while doing literature research.
From a list of input gene names, it fetches abstracts from PubMed, calculates the relationships between genes portrayed in each abstract, then draws a graph displaying these relationships. Graph edges are coloured to indicate stimulatory or inhibitory relationships, and clicking on the edge labels loads the corresponding abstracts in the right-hand frame.
This tool, it seems, helps to draw separate ideas together: if you already know a lot about each gene/protein separately, but need to find connections between them, a graph of this kind is the ideal way to explore that space.